We're wasting money by only supporting gzip for raw DNA files.
- The increasing throughput of Illumina DNA sequencing means institutions and companies are spending tens of thousands of dollars to store terabytes of raw DNA sequence (FASTQ). This data is stored using gzip, a 30-year-old compression algorithm.
- Common bioinformatics tools should support more recent compression algorithms such as zstd for FASTQ data. Zstd has wide industry support, with comparable run times and would likely reduce storage costs by 50% over gzip.
Gzip is outperformed by other algorithms
The original implementation of gzip (Gailly/Madler) has been surpassed in performance by other gzip implementations. For example, cloudflare-zlib outperforms the original gzip in compression speeds and should be used instead.
The use of the gzip compression format is still ubiquitous for raw FASTQ DNA sequence. This is due to it being the only supported compression format for FASTQ in bioinformatics tools. In the thirty years since gzip was created there are now alternatives with superior compression ratios. Only supporting gzip for FASTQ translates into millions of dollars in storage fees on services like Amazon’s S3 and EFS compared with algorithms with better compression ratios. Companies like Meta, Amazon, and Uber are reported to be switching to zstd over gzip. If the most common bioinformatics tools can move to support ingesting zstd-compressed FASTQ format this could save everyone time and money with minimal impact on compression times.
A toy benchmark
As an example a zstd compressed FASTQ file
(SRR7589561)
is almost 50% the size of the same gzipped file. In the figure below I
downloaded ~1.5Gb of FASTQ data and compressed it with either pigz or
zstd. Pigz is a parallel implementation of the original gzip.
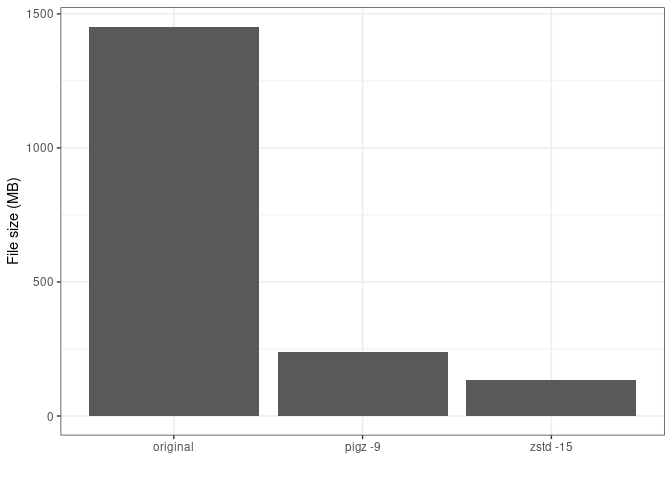
FASTQ file size by compression algorithm.
FASTQ files do however take longer to compress with zstd. The ztsd -15
command takes ~70s which is 100% longer than pigz -9 at ~35s. However,
it’s worth noting when storing raw FASTQ from a sequencer, these files
are compressed once, and then stored for years. This additional CPU time
cost is more than offset by savings in storage costs. The same does not
apply to intermediate files such as trimmed or filtered FASTQ in a
pipeline that tend to be ephemeral. These would require a further
examination of trade offs.

Total compression time in seconds by algorithm.
The next figure shows that changes in decompression time for the same file are relatively small, 3.5s vs 2.2s. Therefore decompression would be minimally impacted.
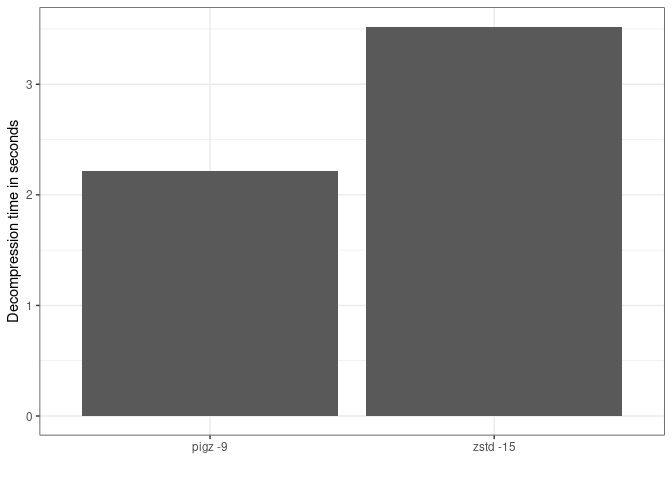
Total decompression time in seconds by algorithm.
Detailed comparison of flags
This figure compares the compressed output file size for all the different available gzip implementations with zstd for different compression flags on the same SRR7589561 FASTQ file. This shows that zstd outperforms gzip at the highest compression levels, with the output file sizes being ~60% the size of the highest gzip compression levels.

Output compressed file size ratios by command line flag for each compression tool. Each colour represents a different compression tool implementation. Each argument was benchmarked five times. Note that zopfli has a single datum because it only compresses to the max ratio.
This next plot compares the trade-offs for file size versus the wall clock run time taken to compress a FASTQ file. This is for the compression process running single-threaded. This shows that zstd can result in much better compression ratios, ~10% of the original file size but with increasing run time. Though not nearly as long as the run time for zopfli, a gzip implementation gives the best compression ratio of any gzip implementation but at the expense ~2 orders of magnitude in compression time.

Compression ratio versus compression time. Each colour represents a different compression tool implementation. Each argument was benchmarked five times. Note that zopfli has a single point because it only compresses to the max ratio.
Takeaway
The gzip implementation is superseded by other compression algorithms such as zstd. By continuing to only support gzip for FASTQ, the bioinformatics industry spends money unnecessarily on additional storage. Bioinformatics tools should widely support zstd as a compression format for FASTQ.
